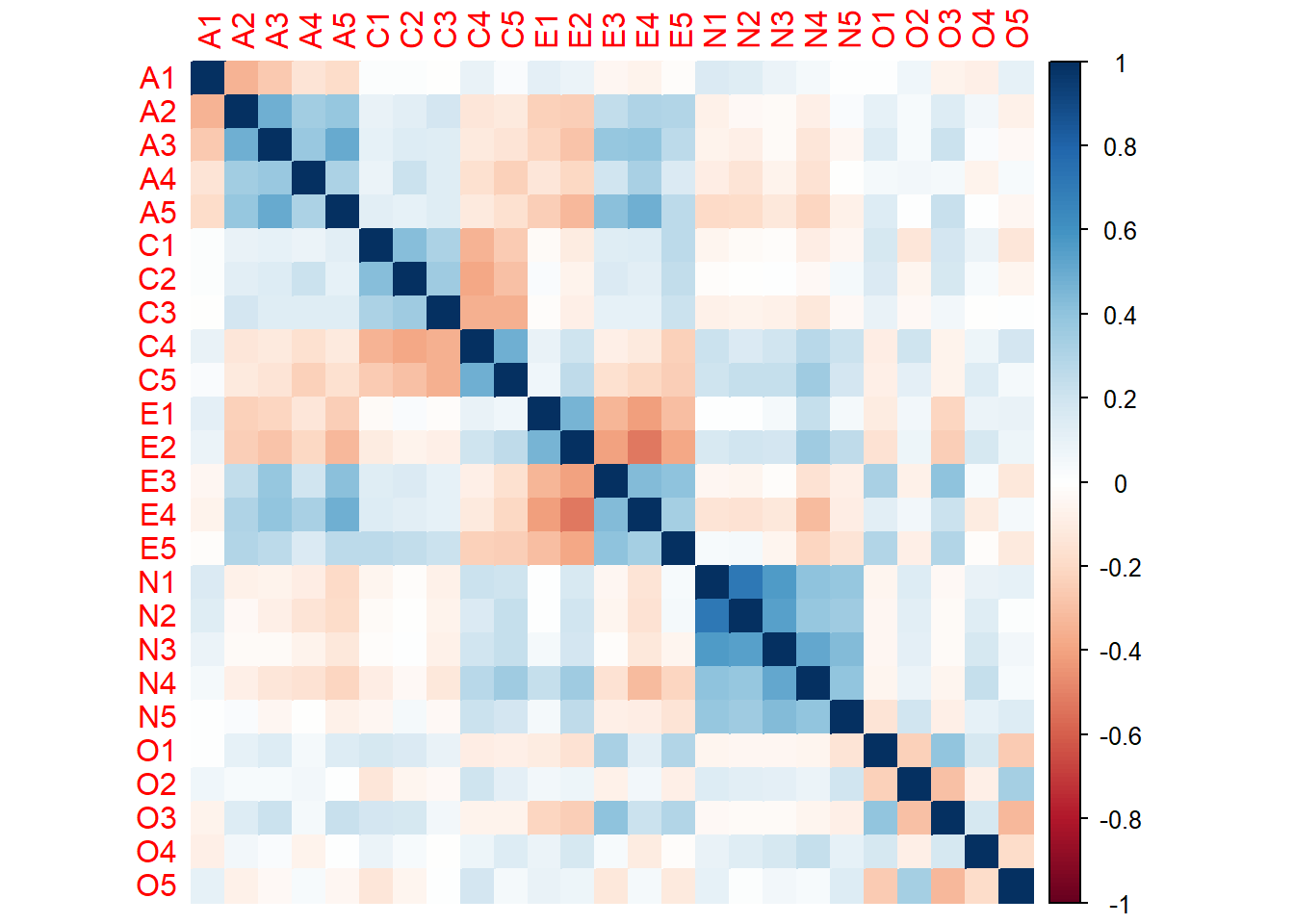
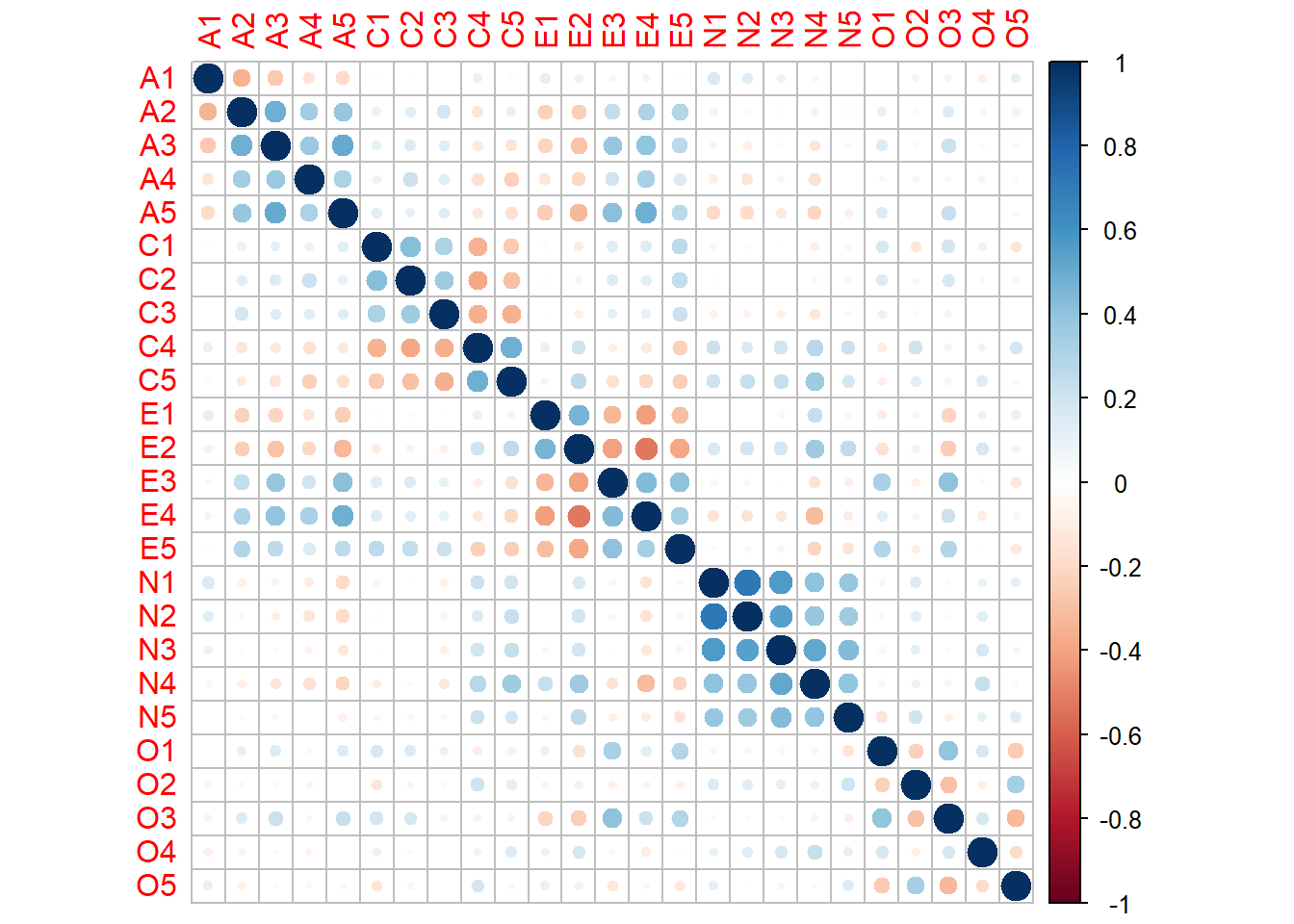
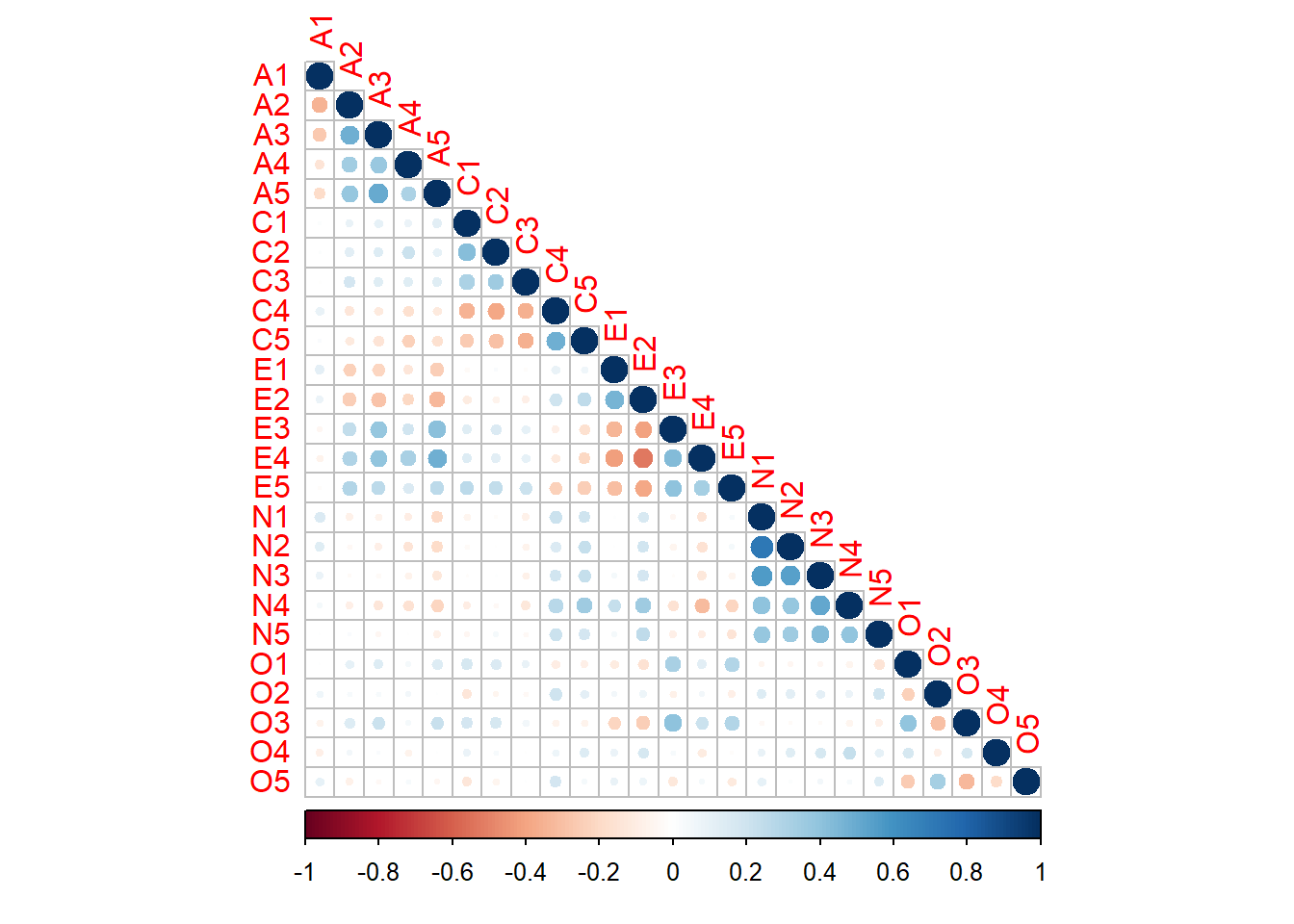
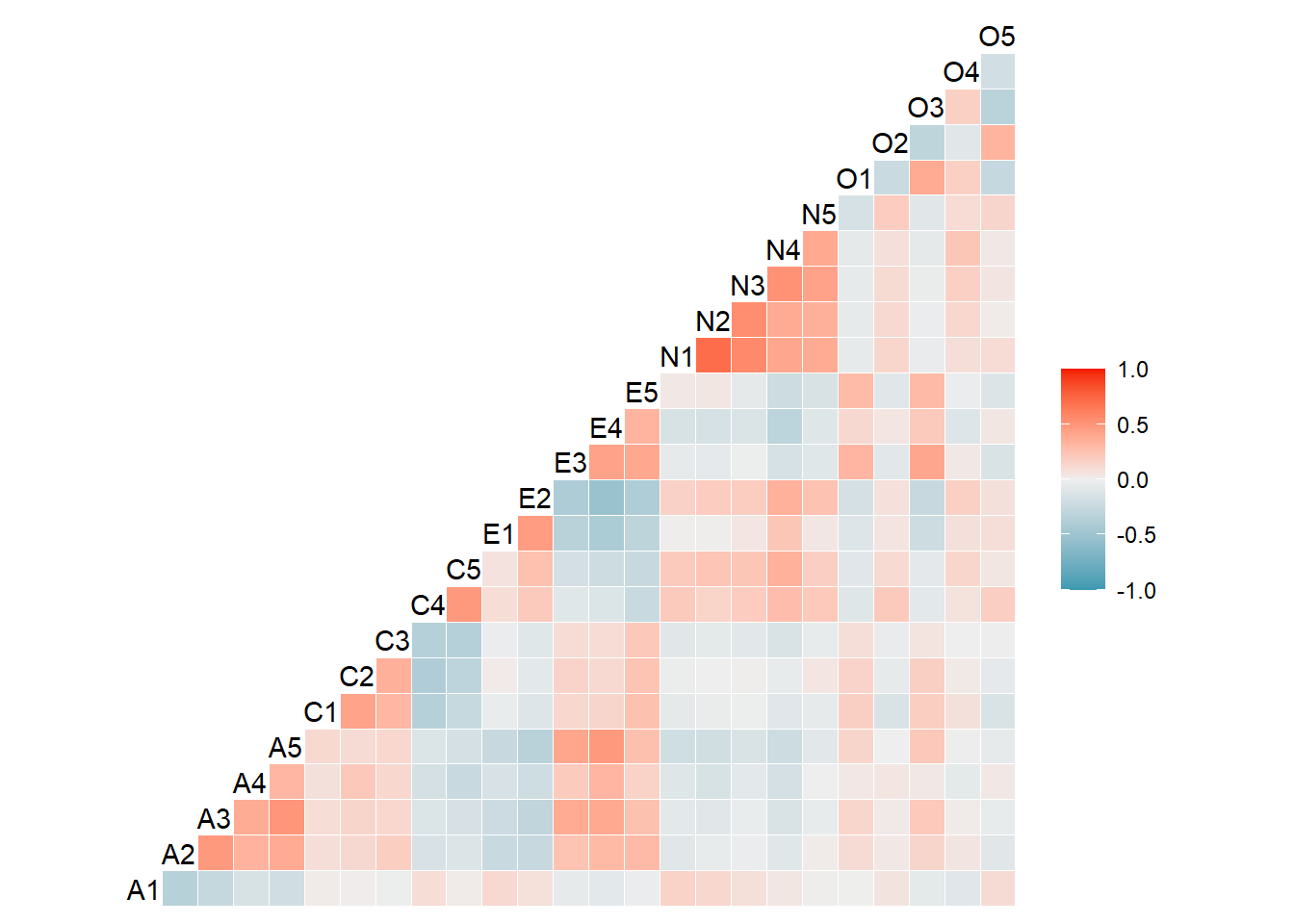
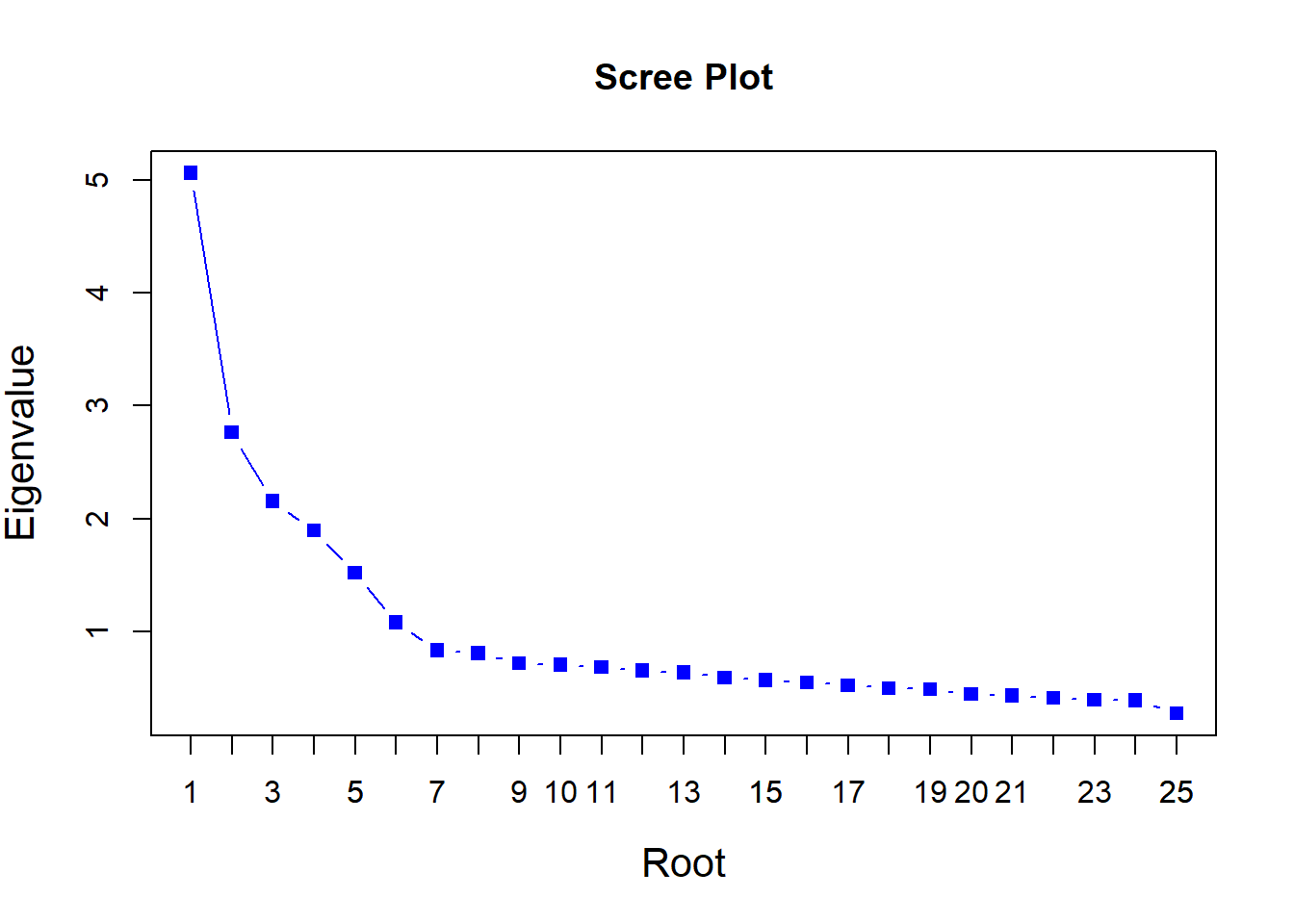
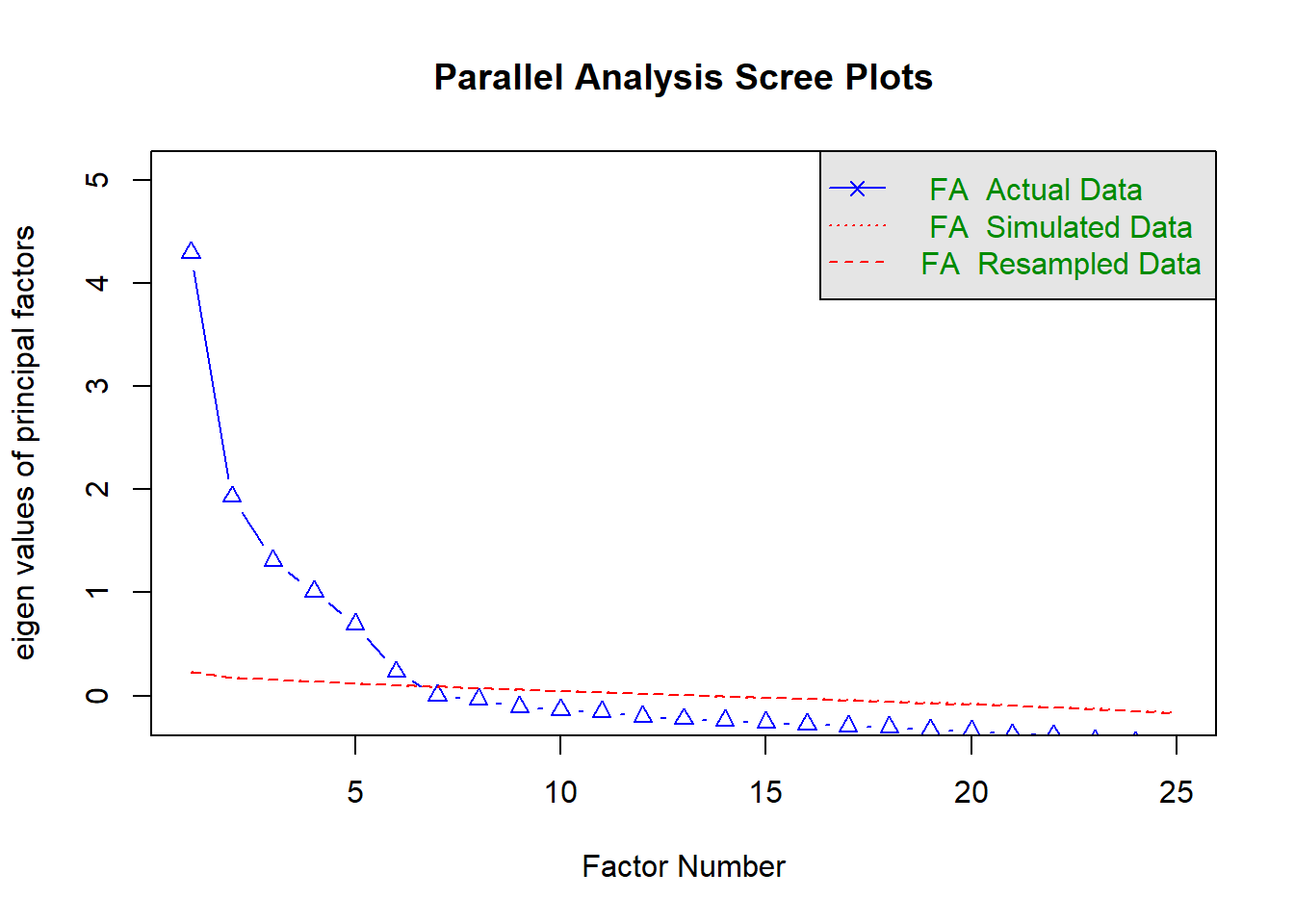
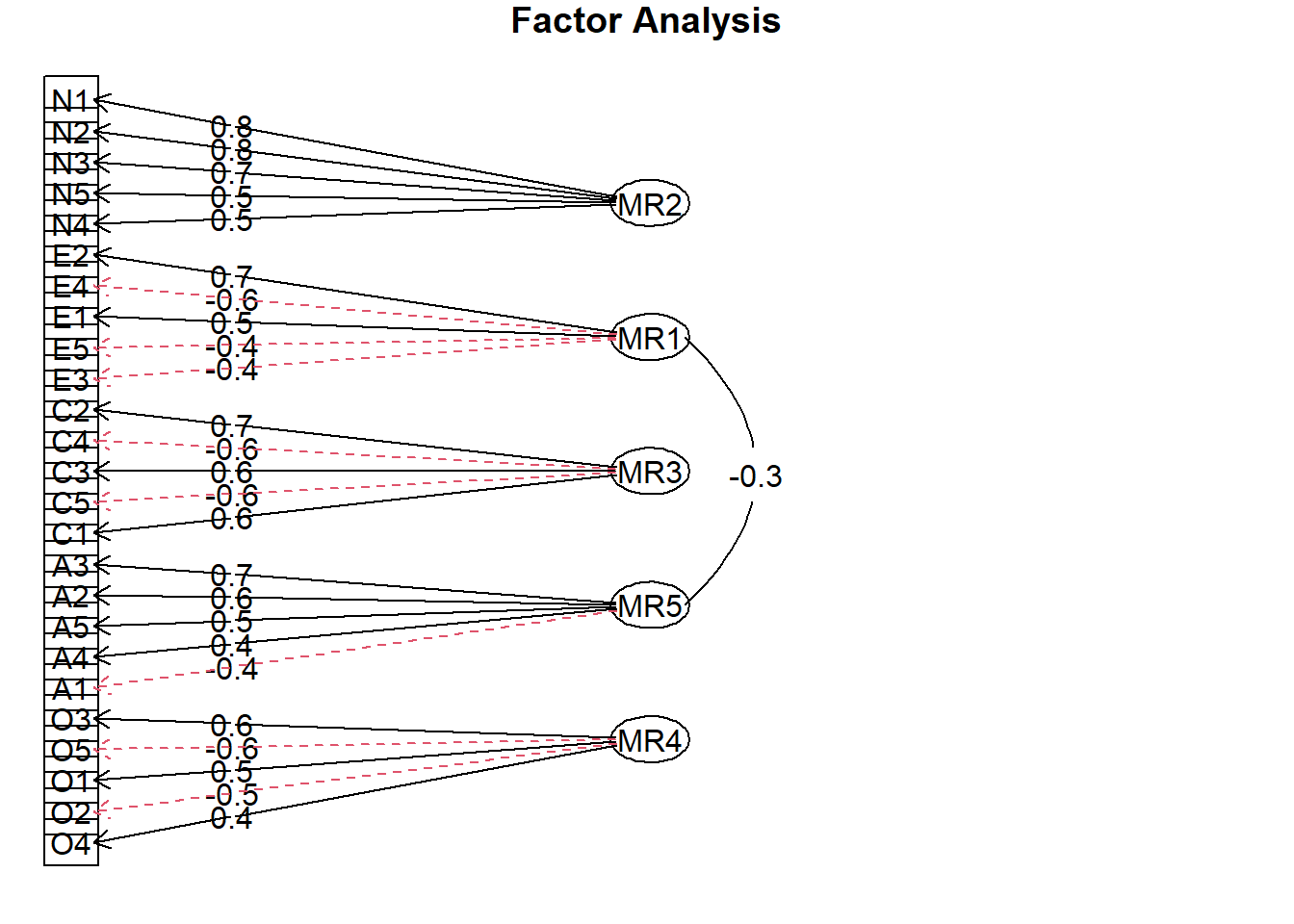
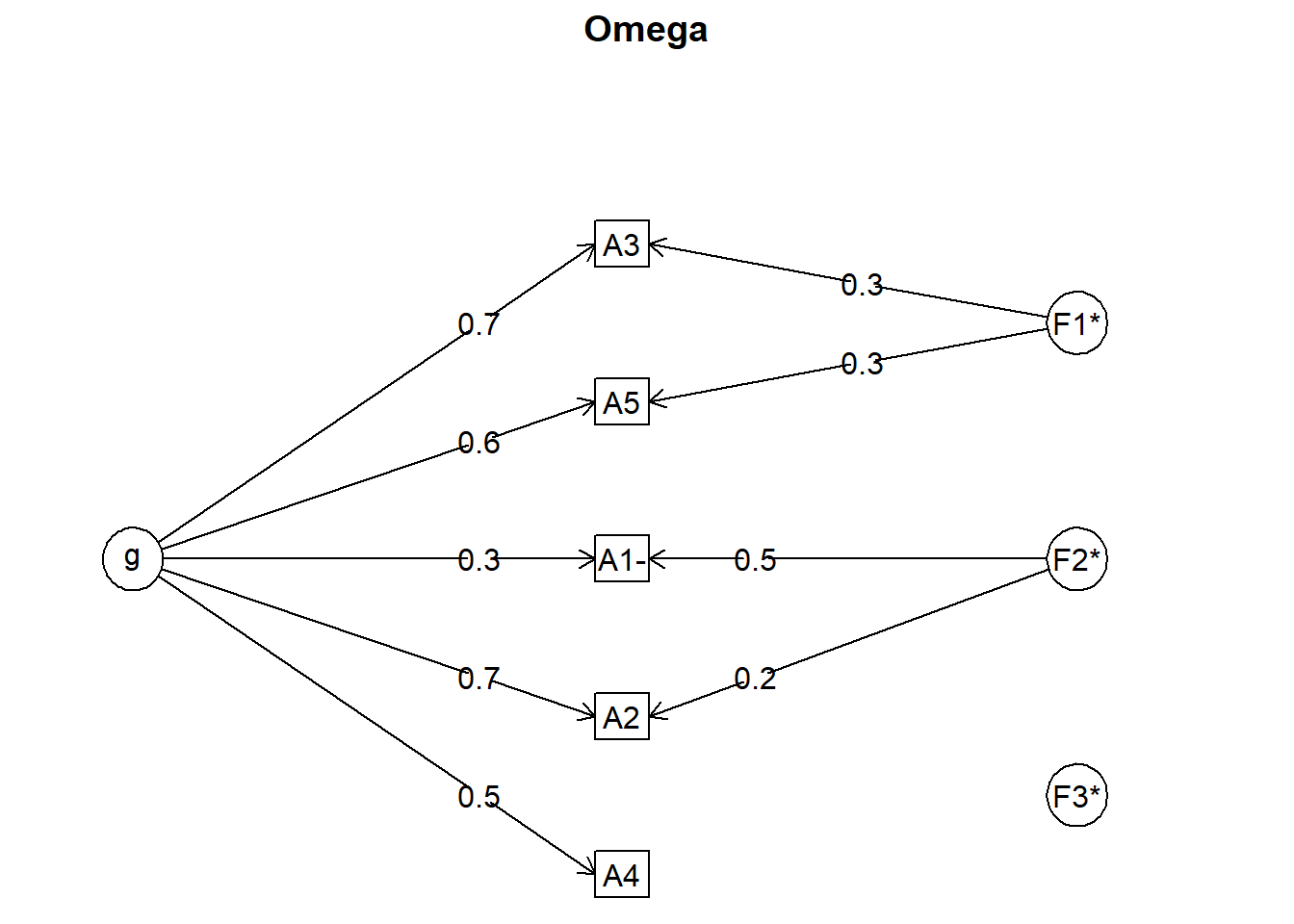
Rows: 2,800
Columns: 28
$ A1 <int> 2, 2, 5, 4, 2, 6, 2, 4, 4, 2, 4, 2, 5, 5, 4, 4, 4, 5, 4, 4, …
$ A2 <int> 4, 4, 4, 4, 3, 6, 5, 3, 3, 5, 4, 5, 5, 5, 5, 3, 6, 5, 4, 4, …
$ A3 <int> 3, 5, 5, 6, 3, 5, 5, 1, 6, 6, 5, 5, 5, 5, 2, 6, 6, 5, 5, 6, …
$ A4 <int> 4, 2, 4, 5, 4, 6, 3, 5, 3, 6, 6, 5, 6, 6, 2, 6, 2, 4, 4, 5, …
$ A5 <int> 4, 5, 4, 5, 5, 5, 5, 1, 3, 5, 5, 5, 4, 6, 1, 3, 5, 5, 3, 5, …
$ C1 <int> 2, 5, 4, 4, 4, 6, 5, 3, 6, 6, 4, 5, 5, 4, 5, 5, 4, 5, 5, 1, …
$ C2 <int> 3, 4, 5, 4, 4, 6, 4, 2, 6, 5, 3, 4, 4, 4, 5, 5, 4, 5, 4, 1, …
$ C3 <int> 3, 4, 4, 3, 5, 6, 4, 4, 3, 6, 5, 5, 3, 4, 5, 5, 4, 5, 5, 1, …
$ C4 <int> 4, 3, 2, 5, 3, 1, 2, 2, 4, 2, 3, 4, 2, 2, 2, 3, 4, 4, 4, 5, …
$ C5 <int> 4, 4, 5, 5, 2, 3, 3, 4, 5, 1, 2, 5, 2, 1, 2, 5, 4, 3, 6, 6, …
$ E1 <int> 3, 1, 2, 5, 2, 2, 4, 3, 5, 2, 1, 3, 3, 2, 3, 1, 1, 2, 1, 1, …
$ E2 <int> 3, 1, 4, 3, 2, 1, 3, 6, 3, 2, 3, 3, 3, 2, 4, 1, 2, 2, 2, 1, …
$ E3 <int> 3, 6, 4, 4, 5, 6, 4, 4, NA, 4, 2, 4, 3, 4, 3, 6, 5, 4, 4, 4,…
$ E4 <int> 4, 4, 4, 4, 4, 5, 5, 2, 4, 5, 5, 5, 2, 6, 6, 6, 5, 6, 5, 5, …
$ E5 <int> 4, 3, 5, 4, 5, 6, 5, 1, 3, 5, 4, 4, 4, 5, 5, 4, 5, 6, 5, 6, …
$ N1 <int> 3, 3, 4, 2, 2, 3, 1, 6, 5, 5, 3, 4, 1, 1, 2, 4, 4, 6, 5, 5, …
$ N2 <int> 4, 3, 5, 5, 3, 5, 2, 3, 5, 5, 3, 5, 2, 1, 4, 5, 4, 5, 6, 5, …
$ N3 <int> 2, 3, 4, 2, 4, 2, 2, 2, 2, 5, 4, 3, 2, 1, 2, 4, 4, 5, 5, 5, …
$ N4 <int> 2, 5, 2, 4, 4, 2, 1, 6, 3, 2, 2, 2, 2, 2, 2, 5, 4, 4, 5, 1, …
$ N5 <int> 3, 5, 3, 1, 3, 3, 1, 4, 3, 4, 3, NA, 2, 1, 3, 5, 5, 4, 2, 1,…
$ O1 <int> 3, 4, 4, 3, 3, 4, 5, 3, 6, 5, 5, 4, 4, 5, 5, 6, 5, 5, 4, 4, …
$ O2 <int> 6, 2, 2, 3, 3, 3, 2, 2, 6, 1, 3, 6, 2, 3, 2, 6, 1, 1, 2, 1, …
$ O3 <int> 3, 4, 5, 4, 4, 5, 5, 4, 6, 5, 5, 4, 4, 4, 5, 6, 5, 4, 2, 5, …
$ O4 <int> 4, 3, 5, 3, 3, 6, 6, 5, 6, 5, 6, 5, 5, 4, 5, 3, 6, 5, 4, 3, …
$ O5 <int> 3, 3, 2, 5, 3, 1, 1, 3, 1, 2, 3, 4, 2, 4, 5, 2, 3, 4, 2, 2, …
$ gender <int> 1, 2, 2, 2, 1, 2, 1, 1, 1, 2, 1, 1, 2, 1, 1, 1, 2, 1, 2, 2, …
$ education <int> NA, NA, NA, NA, NA, 3, NA, 2, 1, NA, 1, NA, NA, NA, 1, NA, N…
$ age <int> 16, 18, 17, 17, 17, 21, 18, 19, 19, 17, 21, 16, 16, 16, 17, …